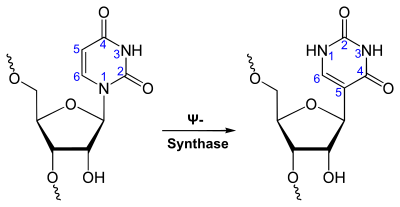Pseudouridine
 | |
| Names | |
|---|---|
IUPAC name 5-[(2S,3R,4S,5R)-3,4-Dihydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-pyrimidine-2,4-dione | |
Preferred IUPAC name 5-(β-D-ribofuranosyl)pyrimidine-2,4(1H,3H)-dione | |
| Other names psi-Uridine, 5-Ribosyluracil, beta-D-Pseudouridine, 5-(beta-D-Ribofuranosyl)uracil | |
| Identifiers | |
CAS Number |
|
3D model (JSmol) |
|
ChEBI |
|
ChemSpider |
|
PubChem CID |
|
InChI
| |
SMILES
| |
| Properties | |
Chemical formula | C9H12N2O6 |
Molar mass | 244.20 g/mol |
| Appearance | White granular powder |
Solubility in water | Highly soluble in water. |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Infobox references | |
Pseudouridine (abbreviated by the Greek letter psi- Ψ or the letter Q)[1] is an isomer of the nucleoside uridine in which the uracil is attached via a carbon-carbon instead of a nitrogen-carbon glycosidic bond. (In this configuration uracil is sometimes referred to as 'pseudouracil'.) It is the most prevalent of the over one hundred different modified nucleosides found in RNA.[2] Ψ is found in all species and in many classes of RNA.[3][4][5][6] Ψ is formed by enzymes called Ψ synthases, which post-transcriptionally isomerize specific uridine residues in RNA in a process termed pseudouridylation.[7] Currently, about ∼ 9500 pseudouridine (Ψ) modifications have been identified in mammals and yeast and deposited in RMBase database.[8]
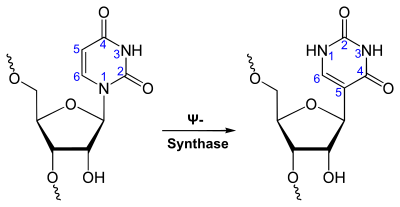
Pseudouridine is biosynthesized from uridine via the action of Ψ synthases.
It is commonly found in tRNA, associated with thymidine and cytosine in the TΨC arm and is one of the invariant regions of tRNA. The function of it is not very clear, but it is expected to play a role in association with aminoacyl transferases during their interaction with tRNA, and hence in the initiation of translation. Recent studies suggest it may offer protection from radiation.[9]

A tRNAAla from S. cerevisiae.
Pseudouridine = Ψ
See also
- Pseudouridine kinase
- TRNA-pseudouridine synthase
References
^ IUPAC-IUB Commission on Biochemical Nomenclature (1970). "Abbreviations and symbols for nucleic acids, polynucleotides, and their constituents". Biochemistry. 9 (20): 4022–4027. doi:10.1021/bi00822a023. PMC 1179624. PMID 5499957..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output .citation q{quotes:"""""""'""'"}.mw-parser-output .citation .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-ws-icon a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-maint{display:none;color:#33aa33;margin-left:0.3em}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Hamma T, Ferré-D'Amaré AR (2006). "Pseudouridine synthases". Chem. Biol. 13 (11): 1125–1135. doi:10.1016/j.chembiol.2006.09.009. PMID 17113994.
^ Charette M, Gray MW (2000). "Pseudouridine in RNA: what, where, how, and why". IUBMB Life. 49 (5): 341–351. doi:10.1080/152165400410182. PMID 10902565.
^ Liang XH, Xu YX, MIchaeli S (2002). "The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA". RNA. 8 (2): 237–246. doi:10.1017/S1355838202018290. PMC 1370245. PMID 11911368.
^ Carlile, Thomas (2014-09-05). "Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells". Nature. 515 (7525): 143–6. doi:10.1038/nature13802. PMC 4224642. PMID 25192136.
^ Schwartz, Schraga (2014-09-11). "Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA". Cell. 159 (1): 148–62. doi:10.1016/j.cell.2014.08.028. PMC 4180118. PMID 25219674.
^ Ferré-D'Amaré AR (2003). "RNA-modifying enzymes". Curr. Opin. Struct. Biol. 13 (1): 49–55. doi:10.1016/S0959-440X(02)00002-7. PMID 12581659.
^ Sun, WJ; Li, JH; Liu, S; Wu, J; Zhou, H; Qu, LH; Yang, JH (4 January 2016). "RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data". Nucleic Acids Research. 44 (D1): D259–65. doi:10.1093/nar/gkv1036. PMC 4702777. PMID 26464443.
^ Monobe, Manami; Arimoto-Kobayashi, Sakae; Ando, Koichi (2003). "β-Pseudouridine, a beer component, reduces radiation-induced chromosome aberrations in human lymphocytes". Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 538 (1–2): 93–99. doi:10.1016/S1383-5718(03)00094-9. PMID 12834758.