Major histocompatibility complex
| Major histocompatibility complex | |
|---|---|
| Identifiers | |
| Symbol | HLA |
| InterPro | IPR001039 |
| Membranome | 63 |
The major histocompatibility complex (MHC) is a set of cell surface proteins essential for the acquired immune system to recognize foreign molecules in vertebrates, which in turn determines histocompatibility. The main function of MHC molecules is to bind to antigens derived from pathogens and display them on the cell surface for recognition by the appropriate T-cells.[1] MHC molecules mediate interactions of leukocytes, also called white blood cells (WBCs), which are immune cells, with other leukocytes or with body cells. The MHC determines compatibility of donors for organ transplant, as well as one's susceptibility to an autoimmune disease via crossreacting immunization. The human MHC is also called the HLA (human leukocyte antigen) complex (often just the HLA). The MHC in mice is called the H-2 complex or H-2.
In a cell, protein molecules of the host's own phenotype or of other biologic entities are continually synthesized and degraded. Each MHC molecule on the cell surface displays a molecular fraction of a protein, called an epitope.[2] The presented antigen can be either self or non-self, thus preventing an organism's immune system targeting its own cells. In its entirety, the MHC population is like a meter indicating the balance of proteins within the cell.
The MHC gene family is divided into three subgroups: class I, class II, and class III. Class I MHC molecules have β2 subunits which can only be recognised by CD8 co-receptors. Class II MHC molecules have β1 and β2 subunits and can be recognised by CD4 co-receptors. In this way MHC molecules chaperone which type of lymphocytes may bind to the given antigen with high affinity, since different lymphocytes express different T-Cell Receptor (TCR) co-receptors.
Diversity of antigen presentation, mediated by MHC classes I and II, is attained in at least three ways: (1) an organism's MHC repertoire is polygenic (via multiple, interacting genes); (2) MHC expression is codominant (from both sets of inherited alleles); (3) MHC gene variants are highly polymorphic (diversely varying from organism to organism within a species).[3]Major histocompatibility complex and sexual selection has been observed in male mice making mate choices of females with different MHCs and thus demonstrating sexual selection.[4] Also, at least for MHC I presentation, there has been evidence of antigenic peptide splicing which can combine peptides from different proteins, vastly increasing antigen diversity.[5]
Contents
1 Discovery
2 In immunity
2.1 Functions
2.2 Lymphocytes
2.3 MHC class II
2.4 MHC class I
3 Genes
4 Proteins
4.1 Class I
4.2 Class II
4.3 Class III
5 Antigen processing and presentation
6 T lymphocyte recognition restrictions
7 In sexual mate selection
8 Evolutionary diversity
9 In transplant rejection
10 HLA biology
11 See also
12 Notes and references
13 Bibliography
14 External links
Discovery
The first descriptions of the MHC were made by British immunologist Peter Gorer in 1936.[6] MHC genes were first identified in inbred mice strains. Clarence Little transplanted tumors across differing strains and found rejection of transplanted tumors according to strains of host versus donor.[7]George Snell selectively bred two mouse strains, attained a new strain nearly identical to one of the progenitor strains, but differing crucially in histocompatibility—that is, tissue compatibility upon transplantation—and thereupon identified an MHC locus.[8] For this work, Snell was awarded the 1980 Nobel Prize in Physiology or Medicine, together with Baruj Benacerraf and Jean Dausset.
In immunity
Of the three MHC classes identified, attention commonly focuses on classes I and II. By interacting with CD4 molecules on surfaces of helper T cells, MHC class II mediates establishment of specific immunity (also called acquired immunity or adaptive immunity). By interacting with CD8 molecules on surfaces of cytotoxic T cells, MHC class I mediates destruction of infected or malignant host cells, the aspect of specific immunity termed cellular immunity. (The other arm of specific immunity is humoral immunity, whose relation to MHC is more indirect.)
Functions
MHC is the tissue-antigen that allows the immune system (more specifically T cells) to bind to, recognize, and tolerate itself (autorecognition). MHC is also the chaperone for intracellular peptides that are complexed with MHCs and presented to T cell receptors (TCRs) as potential foreign antigens. MHC interacts with TCR and its co-receptors to optimize binding conditions for the TCR-antigen interaction, in terms of antigen binding affinity and specificity, and signal transduction effectiveness.
Essentially, the MHC-peptide complex is a complex of autoantigen/alloantigen. Upon binding, T cells should in principle tolerate the auto-antigen, but activate when exposed to the allo-antigen. Disease states occur when this principle is disrupted.
Antigen presentation: MHC molecules bind to both T cell receptor and CD4/CD8 co-receptors on T lymphocytes, and the antigen epitope held in the peptide-binding groove of the MHC molecule interacts with the variable Ig-Like domain of the TCR to trigger T-cell activation[9]
Autoimmune reaction: Having some MHC molecules increases the risk of autoimmune diseases more than having others. HLA-B27 is an example. It is unclear how exactly having the HLA-B27 tissue type increases the risk of ankylosing spondylitis and other associated inflammatory diseases, but mechanisms involving aberrant antigen presentation or T cell activation have been hypothesized.
Tissue allorecognition: MHC molecules in complex with peptide epitopes are essentially ligands for TCR. T cells become activated by binding to the peptide-binding grooves of any MHC molecule that T cells were not trained to recognize during thymus positive selection.
Lymphocytes
As a lineage of leukocytes, lymphocytes reside in peripheral lymphoid tissues, including lymphoid follicles and lymph nodes, and include B cells, T cells, and natural killer cells (NK cells). B cells, which act specifically, secrete antibody molecules, but do not bind MHC. T cells, which act specifically, as well as NK cells, which act innately, interact with MHC. NK cells express Killer Ig-like receptors (KIRs) that bind to MHC I molecules and signal through ITIM (immunoreceptor tyrosine inhibition motif) recruitment and activation of protein tyrosine phosphatases. This means in contrast to CD8/TCR interaction that activates Tc lymphocytes, NK cells becomes deactivated when bound to MHC I. When MHC class I expression is low, as is typically the case with abnormal cell function during viral infection or tumourigenesis, NK cells lose the inhibitory KIR signal and trigger programmed cell death of the abnormal cell. NK cells thus help prevent progress of cancerous cells by contributing to tumor surveillance.
MHC class II
MHC class II can be conditionally expressed by all cell types, but normally occurs only on professional antigen-presenting cells (APCs): macrophages, B cells, and especially dendritic cells (DCs). An APC takes up an antigenic protein, performs antigen processing, and returns a molecular fraction of it—a fraction termed the epitope—and displays it on the APC's surface coupled within an MHC class II molecule (antigen presentation). On the cell's surface, the epitope can be recognized by immunologic structures like T cell receptors (TCRs). The molecular region which binds to the epitope is the paratope.
On surfaces of helper T cells are CD4 receptors, as well as TCRs. When a naive helper T cell's CD4 molecule docks to an APC's MHC class II molecule, its TCR can meet and be imprinted by the epitope coupled within the MHC class II. This event primes the naive helper T cell. According to the local milieu, that is, the balance of cytokines secreted by APCs in the microenvironment, the naive helper T cell (Th0) polarizes into either a memory Th cell or an effector Th cell of phenotype either type 1 (Th1), type 2 (Th2), type 17 (Th17), or regulatory/suppressor (Treg), as so far identified, the Th cell's terminal differentiation.
MHC class II thus mediates immunization to—or, if APCs polarize Th0 cells principally to Treg cells, immune tolerance of—an antigen. The polarization during primary exposure to an antigen is key in determining a number chronic diseases, such as inflammatory bowel diseases and asthma, by skewing the immune response that memory Th cells coordinate when their memory recall is triggered upon secondary exposure to similar antigens. (B cells express MHC class II to present antigen to Th0, but when their B cell receptors bind matching epitopes, interactions which are not mediated by MHC, these activated B cells secrete soluble immunoglobulins: antibody molecules mediating humoral immunity.)
MHC class I
MHC class I occurs on all nucleated cells and also in platelets—in essence all cells but red blood cells. It presents epitopes to killer T cells, also called cytotoxic T lymphocytes (CTLs). A CTL expresses CD8 receptors, in addition to TCRs. When a CTL's CD8 receptor docks to a MHC class I molecule, if the CTL's TCR fits the epitope within the MHC class I molecule, the CTL triggers the cell to undergo programmed cell death by apoptosis. Thus, MHC class I helps mediate cellular immunity, a primary means to address intracellular pathogens, such as viruses and some bacteria, including bacterial L forms, bacterial genus Mycoplasma, and bacterial genus Rickettsia. In humans, MHC class I comprises HLA-A, HLA-B, and HLA-C molecules.
Genes
MHC gene families are found in all vertebrates, though they vary widely. In humans, the MHC region occurs on chromosome 6, between the flanking genetic markers MOG and COL11A2 (from 6p22.1 to 6p21.3 about 29Mb to 33Mb on the hg38 assembly), and contains 224 genes spanning 3.6 megabase pairs (3 600 000 bases).[10] About half have known immune functions.
The same markers in the gray short-tailed opossum (Monodelphis domestica), a marsupial, span 3.95 Mb, yielding 114 genes, 87 shared with humans.[11] Marsupial MHC genotypic variation lies between eutherian mammals and birds, taken as minimal MHC encoding, but is closer in organization to that of nonmammals, and MHC class I genes of marsupials have amplified within the class II region, yielding a unique class I/II region.[11]
Class III functions very differently from class I and class II, but its locus occurs between the other two classes, on chromosome 6 in humans, and are frequently discussed together.
| Class | Encoding | Expression |
| I | (1) peptide-binding proteins, which select short sequences of amino acids for antigen presentation, as well as (2) molecules aiding antigen-processing (such as TAP and tapasin). | One chain, called α, whose ligands are the CD8 receptor—borne notably by cytotoxic T cells—and inhibitory receptors borne by NK cells |
| II | (1) peptide-binding proteins and (2) proteins assisting antigen loading onto MHC class II's peptide-binding proteins (such as MHC II DM, MHC II DQ, MHC II DR, and MHC II DP). | Two chains, called α & β, whose ligands are the CD4 receptors borne by helper T cells. |
| III | Other immune proteins, outside antigen processing and presentation, such as components of the complement cascade (e.g., C2, C4, factor B), the cytokines of immune signaling (e.g., TNF-α), and heat shock proteins buffering cells from stresses | Various |
Proteins
MHC proteins have immunoglobulin-like structure.
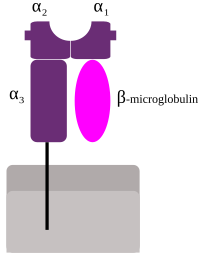
MHC class I protein molecule
Class I
MHC I occurs as an α chain composed of three domains—α1, α2, and α3. The α1 rests upon a unit of the non-MHC molecule β2 microglobulin (encoded on human chromosome 15). The α3 domain is transmembrane, anchoring the MHC class I molecule to the cell membrane. The peptide being presented is held by the floor of the peptide-binding groove, in the central region of the α1/α2 heterodimer (a molecule composed of two nonidentical subunits). The genetically encoded and expressed sequence of amino acids, the sequence of residues, of the peptide-binding groove's floor determines which particular peptide residues it binds.[12]

MHC class II protein molecule
Classical MHC molecules present epitopes to the TCRs of CD8+ T lymphocytes. Nonclassical molecules (MHC class IB) exhibit limited polymorphism, expression patterns, and presented antigens; this group is subdivided into a group encoded within MHC loci (e.g., HLA-E, -F, -G), as well as those not (e.g., stress ligands such as ULBPs, Rae1, and H60); the antigen/ligand for many of these molecules remain unknown, but they can interact with each of CD8+ T cells, NKT cells, and NK cells.
Class II
MHC class II is formed of two chains, α and β, each having two domains—α1 and α2 and β1 and β2—each chain having a transmembrane domain, α2 and β2, respectively, anchoring the MHC class II molecule to the cell membrane.[13] The peptide-binding groove is formed of the heterodimer of α1 and β1.
MHC class II molecules in humans have five to six isotypes. Classic molecules present peptides to CD4+ lymphocytes. Nonclassic molecules, accessories, with intracellular functions, are not exposed on cell membranes, but in internal membranes in lysosomes, normally loading the antigenic peptides onto classic MHC class II molecules.
Class III
Class III molecules have physiologic roles unlike classes I and II, but are encoded between them in the short arm of human chromosome 6. Class III molecules include several secreted proteins with immune functions: components of the complement system (such as C2, C4, and B factor), cytokines (such as TNF-α, LTA, and LTB), and heat shock proteins.
Antigen processing and presentation

MHC class I pathway: Proteins in the cytosol are degraded by the proteasome, liberating peptides internalized by TAP channel in the endoplasmic reticulum, there associating with MHC-I molecules freshly synthesized. MHC-I/peptide complexes enter Golgi apparatus, are glycosylated, enter secretory vesicles, fuse with the cell membrane, and externalize on the cell membrane interacting with T lymphocytes.
Peptides are processed and presented by two classical pathways:
- In MHC class II, phagocytes such as macrophages and immature dendritic cells take up entities by phagocytosis into phagosomes—though B cells exhibit the more general endocytosis into endosomes—which fuse with lysosomes whose acidic enzymes cleave the uptaken protein into many different peptides. Via physicochemical dynamics in molecular interaction with the particular MHC class II variants borne by the host, encoded in the host's genome, a particular peptide exhibits immunodominance and loads onto MHC class II molecules. These are trafficked to and externalized on the cell surface.[14]
- In MHC class I, any nucleated cell normally presents cytosolic peptides, mostly self peptides derived from protein turnover and defective ribosomal products. During viral infection, intracellular microorganism infection, or cancerous transformation, such proteins degraded in the proteosome are as well loaded onto MHC class I molecules and displayed on the cell surface. T lymphocytes can detect a peptide displayed at 0.1%-1% of the MHC molecules.

Peptide binding for Class I and Class II MHC molecules, showing the binding of peptides between the alpha-helix walls, upon a beta-sheet base. The difference in binding positions is shown. Class I primarily makes contact with backbone residues at the Carboxy and amino terminal regions, while Class II primarily makes contacts along the length of the residue backbone. The precise location of binding residues is determined by the MHC allele.[15]
| Characteristic | MHC-I pathway | MHC-II pathway |
|---|---|---|
| Composition of the stable peptide-MHC complex | Polymorphic chain α and β2 microglobulin, peptide bound to α chain | Polymorphic chains α and β, peptide binds to both |
| Types of antigen presenting cells (APC) | All nucleated cells | Dendritic cells, mononuclear phagocytes, B lymphocytes, some endothelial cells, epithelium of thymus |
| T lymphocytes able to respond | Cytotoxic T lymphocytes (CD8+) | Helper T lymphocytes (CD4+) |
| Origin of antigenic proteins | cytosolic proteins (mostly synthetized by the cell; may also enter from the extracellular medium via phagosomes) | Proteins present in endosomes or lysosomes (mostly internalized from extracellular medium) |
| Enzymes responsible for peptide generation | Cytosolic proteasome | Proteases from endosomes and lysosomes (for instance, cathepsin) |
| Location of loading the peptide on the MHC molecule | Endoplasmic reticulum | Specialized vesicular compartment |
| Molecules implicated in transporting the peptides and loading them on the MHC molecules | TAP (transporter associated with antigen processing) | DM, invariant chain |
T lymphocyte recognition restrictions
In their development in the thymus, T lymphocytes are selected to recognize MHC molecules of the host, but not recognize other self antigens. Following selection, each T lymphocyte shows dual specificity: The TCR recognizes self MHC, but only non-self antigens.
MHC restriction occurs during lymphocyte development in the thymus through a process known as positive selection. T cells that do not receive a positive survival signal — mediated mainly by thymic epithelial cells presenting self peptides bound to MHC molecules — to their TCR undergo apoptosis. Positive selection ensures that mature T cells can functionally recognize MHC molecules in the periphery (i.e. elsewhere in the body).
The TCRs of T lymphocytes recognise only sequential epitopes, also called linear epitopes, of only peptides and only if coupled within an MHC molecule. (Antibody molecules secreted by activated B cells, though, ligate diverse epitopes—peptide, lipid, carbohydrate, and nucleic acid—and recognize conformational epitopes, which have three-dimensional structure.)
In sexual mate selection
MHC molecules enable immune system surveillance of the population of protein molecules in a host cell, and greater MHC diversity permits greater diversity of antigen presentation. In 1976, Yamazaki et al demonstrated a sexual selection mate choice by male mice for females of a different MHC. Similar results have been obtained with fish.[16] Some data find lower rates of early pregnancy loss in human couples of dissimilar MHC genes.[17]
MHC may be related to mate choice in some human populations, a theory that found support by studies by Ober and colleagues in 1997,[18] as well as by Chaix and colleagues in 2008.[19] However, the latter findings have been controversial.[20] If it exists, the phenomenon might be mediated by olfaction, as MHC phenotype appears strongly involved in the strength and pleasantness of perceived odour of compounds from sweat. Fatty acid esters—such as methyl undecanoate, methyl decanoate, methyl nonanoate, methyl octanoate, and methyl hexanoate—show strong connection to MHC.[21]
In 1995, Claus Wedekind found that in a group of female college students who smelled T-shirts worn by male students for two nights (without deodorant, cologne, or scented soaps), by far most women chose shirts worn by men of dissimilar MHCs, a preference reversed if the women were on oral contraceptives.[22] Results of a 2002 experiment likewise suggest HLA-associated odors influence odor preference and may mediate social cues.[23] In 2005 in a group of 58 subjects, women were more indecisive when presented with MHCs like their own,[24] although with oral contraceptives, the women showed no particular preference.[25] No studies show the extent to which odor preference determines mate selection (or vice versa).
Evolutionary diversity
Most mammals have MHC variants similar to those of humans, who bear great allelic diversity, especially among the nine classical genes—seemingly due largely to gene duplication—though human MHC regions have many pseudogenes. The most diverse loci, namely HLA-A, HLA-B, and HLA-DRB1, have roughly 1000, 1600, and 870 known alleles, respectively[citation needed].[26] Many HLA alleles are ancient, sometimes of closer homology to a chimpanzee MHC alleles than to some other human alleles of the same gene.
MHC allelic diversity has challenged evolutionary biologists for explanation. Most posit balancing selection (see polymorphism (biology)), which is any natural selection process whereby no single allele is absolutely most fit, such as frequency-dependent selection and heterozygote advantage. Recent models suggest a high number of alleles is implausible via heterozygote advantage alone.[citation needed]
Pathogenic coevolution, a counterhypothesis, posits that common alleles are under greatest pathogenic pressure, driving positive selection of uncommon alleles—moving targets, so to say, for pathogens. As pathogenic pressure on the previously common alleles decreases, their frequency in the population stabilizes, and remain circulating in a large population. Despite great MHC polymorphism at the population level, an individual bears at most 18 MHC I or II alleles.
Relatively low MHC diversity has been observed in the cheetah (Acinonyx jubatus),[27]Eurasian beaver (Castor fiber),[28] and giant panda (Ailuropoda melanoleuca).[29] In 2007 low MHC diversity was attributed a role in disease susceptibility in the Tasmanian devil (Sarcophilus harrisii), native to the isolated island of Tasmania, such that an antigen of a transmissible tumor, involved in devil facial tumour disease, appears to be recognized as a self antigen.[30] To offset inbreeding, efforts to sustain genetic diversity in populations of endangered species and of captive animals have been suggested.
In transplant rejection
In a transplant procedure, as of an organ or stem cells, MHC molecules act themselves as antigens and can provoke immune response in the recipient, thus causing transplant rejection. MHC molecules were identified and named after their role in transplant rejection between mice of different strains, though it took over 20 years to clarify MHC's role in presenting peptide antigens to cytotoxic T lymphocytes (CTLs).[31]
Each human cell expresses six MHC class I alleles (one HLA-A, -B, and -C allele from each parent) and six to eight MHC class II alleles (one HLA-DP and -DQ, and one or two HLA-DR from each parent, and combinations of these). The MHC variation in the human population is high, at least 350 alleles for HLA-A genes, 620 alleles for HLA-B, 400 alleles for DR, and 90 alleles for DQ. Any two individuals who are not identical twins will express differing MHC molecules. All MHC molecules can mediate transplant rejection, but HLA-C and HLA-DP, showing low polymorphism, seem least important.[clarification needed]
When maturing in the thymus, T lymphocytes are selected for their TCR incapacity to recognize self antigens, yet T lymphocytes can react against the donor MHC's peptide-binding groove, the variable region of MHC holding the presented antigen's epitope for recognition by TCR, the matching paratope. T lymphocytes of the recipient take the incompatible peptide-binding groove as nonself antigen. The T lymphocytes' recognition of the foreign MHC as self is allorecognition.[clarification needed]
Transplant rejection has various types known to be mediated by MHC (HLA):
Hyperacute rejection occurs when, before the transplantation, the recipient has preformed anti-HLA antibodies, perhaps by previous blood transfusions (donor tissue that includes lymphocytes expressing HLA molecules), by anti-HLA generated during pregnancy (directed at the father's HLA displayed by the fetus), or by previous transplantation;
Acute cellular rejection occurs when the recipient's T lymphocytes are activated by the donor tissue, causing damage via mechanisms such as direct cytotoxicity from CD8 cells.
Acute humoral rejection and chronic disfunction occurs when the recipient's anti-HLA antibodies form directed at HLA molecules present on endothelial cells of the transplanted tissue.
In all of the above situations, immunity is directed at the transplanted organ, sustaining lesions. A cross-reaction test between potential donor cells and recipient serum seeks to detect presence of preformed anti-HLA antibodies in the potential recipient that recognize donor HLA molecules, so as to prevent hyperacute rejection. In normal circumstances, compatibility between HLA-A, -B, and -DR molecules is assessed. The higher the number of incompatibilities, the lower the five-year survival rate. Global databases of donor information enhance the search for compatible donors.
HLA biology

Codominant expression of HLA genes
Human MHC class I and II are also called human leukocyte antigen (HLA). To clarify the usage, some of the biomedical literature uses HLA to refer specifically to the HLA protein molecules and reserves MHC for the region of the genome that encodes for this molecule, but this is not a consistent convention.
The most studied HLA genes are the nine classical MHC genes: HLA-A, HLA-B, HLA-C, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, and HLA-DRB1. In humans, the MHC gene cluster is divided into three regions: classes I, II, and III. The A, B and C genes belong to MHC class I, whereas the six D genes belong to class II.
MHC alleles are expressed in codominant fashion.[13] This means the alleles (variants) inherited from both parents are expressed equally:
- Each person carries 2 alleles of each of the 3 class-I genes, (HLA-A, HLA-B and HLA-C), and so can express six different types of MHC-I (see figure).
- In the class-II locus, each person inherits a pair of HLA-DP genes (DPA1 and DPB1, which encode α and β chains), a couple of genes HLA-DQ (DQA1 and DQB1, for α and β chains), one gene HLA-DRα (DRA1), and one or more genes HLA-DRβ (DRB1 and DRB3, -4 or -5). That means that one heterozygous individual can inherit six or eight functioning class-II alleles, three or more from each parent. The role of DQA2 or DQB2 is not verified. The DRB2, DRB6, DRB7, DRB8 and DRB9 are pseudogenes.
The set of alleles that is present in each chromosome is called the MHC haplotype. In humans, each HLA allele is named with a number. For instance, for a given individual, his haplotype might be HLA-A2, HLA-B5, HLA-DR3, etc... Each heterozygous individual will have two MHC haplotypes, one each from the paternal and maternal chromosomes.
The MHC genes are highly polymorphic; many different alleles exist in the different individuals inside a population. The polymorphism is so high, in a mixed population (nonendogamic), no two individuals have exactly the same set of MHC molecules, with the exception of identical twins.
The polymorphic regions in each allele are located in the region for peptide contact. Of all the peptides that could be displayed by MHC, only a subset will bind strongly enough to any given HLA allele, so by carrying two alleles for each gene, a much larger set of peptides can be presented.[why?]
On the other hand, inside a population, the presence of many different alleles ensures there will always be an individual with a specific MHC molecule able to load the correct peptide to recognize a specific microbe. The evolution of the MHC polymorphism ensures that a population will not succumb to a new pathogen or a mutated one, because at least some individuals will be able to develop an adequate immune response to win over the pathogen. The variations in the MHC molecules (responsible for the polymorphism) are the result of the inheritance of different MHC molecules, and they are not induced by recombination, as it is the case for the antigen receptors.
Because of the high levels of allelic diversity found within its genes, MHC has also attracted the attention of many evolutionary biologists.[32]
See also
- Cell-mediated immunity
- Disassortative sexual selection
- Humoral immunity
- MHC multimer
- Pheromone
- Streptamer
- Transplant rejection
Notes and references
^ Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001 The Major Histocompatibility Complex and Its Functions
^ Kimball's Biology Histocompatibility Molecules
^ Janeway CA Jr, Travers P, Walport M, et al, Immunobiology: The Immune System in Health and Disease, 5th edn (New York: Garland Science, 2001), "The major histocompatibility complex and its functions".
^ Yamazaki, K.; Boyse, E. A.; Miké, V.; Thaler, H. T.; Mathieson, B. J.; Abbott, J.; Boyse, J.; Zayas, Z. A.; Thomas, L. (1976-11-02). "Control of mating preferences in mice by genes in the major histocompatibility complex". Journal of Experimental Medicine. 144 (5): 1324–1335. doi:10.1084/jem.144.5.1324. ISSN 0022-1007. PMC 2190468. PMID 1032893..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ Vigneron, Nathalie; Stroobant, Vincent; Chapiro, Jacques; Ooms, Annie; Degiovanni, Gérard; Morel, Sandra; Bruggen, Pierre van der; Boon, Thierry; Eynde, Benoît J. Van den (2004-04-23). "An Antigenic Peptide Produced by Peptide Splicing in the Proteasome". Science. 304 (5670): 587–590. doi:10.1126/science.1095522. ISSN 0036-8075. PMID 15001714.
^ Klein J (1986). "Seeds of time: Fifty years ago Peter A. Gorer discovered the H-2 complex". Immunogenetics. 24: 331–338. doi:10.1007/bf00377947.
^ Little CC 1941, "The genetics of tumor transplantation", pp 279–309, in Biology of the Laboratory Mouse, ed by Snell GD, New York: Dover.
^ Snell GD, Higgins GF (1951). "Alleles at the histocompatibility-2 locus in the mouse as determined by tumor transplantation". Genetics. 36: 306–310.
^ Thomas J. Kindt; Richard A. Goldsby; Barbara Anne Osborne; Janis Kuby (2007). Kuby immunology. Macmillan. ISBN 978-1-4292-0211-4. Retrieved 28 November 2010.
^ MHC Sequencing Consortium (1999). "Complete sequence and gene map of a human major histocompatibility complex". Nature. 401 (6756): 921–923. doi:10.1038/44853. PMID 10553908.
^ ab Belov K, Deakin JE, Papenfuss AT, Baker ML, Melman SD, Siddle HV, Gouin N, Goode DL, Sargeant TJ, Robinson MD, Wakefield MJ, Mahony S, Cross JG, Benos PV, Samollow PB, Speed TP, Graves JA, Miller RD (March 2006). "Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex". PLoS Biol. 4 (3): e46. doi:10.1371/journal.pbio.0040046. PMC 1351924. PMID 16435885.
^ Toh H, Savoie CJ, Kamikawaji N, Muta S, Sasazuki T, Kuhara S (October 2000). "Changes at the floor of the peptide-binding groove induce a strong preference for proline at position 3 of the bound peptide: molecular dynamics simulations of HLA-A*0217". Biopolymers. 54 (5): 318–27. doi:10.1002/1097-0282(20001015)54:5<318::AID-BIP30>3.0.CO;2-T. PMID 10935972.
^ ab Abbas; Lichtman A.H. (2009). "Ch.3 Antigen capture and presentation to lymphocytes". Basic Immunology. Functions and disorders of the immune system (3rd ed.). p. A.B. ISBN 978-1-4160-4688-2.
^ Aderem A, Underhill DM (1999). "Mechanisms of phagocytosis in macrophages". Annu. Rev. Immunol. 17: 593–623. doi:10.1146/annurev.immunol.17.1.593. PMID 10358769.
^ K. Murphy, “Antigen recognition by T cells,” in Janeway's Immunobiology, 8th, Ed., Garland Science, 2012, pp. 138-153.
^ Boehm T; Zufall F (2006). "MHC peptides and the sensory evaluation of genotype". Trends Neurosci. 29 (2): 100–107. doi:10.1016/j.tins.2005.11.006. PMID 16337283.
^ Haig D (November 1997). "Maternal-fetal interactions and MHC polymorphism". J. Reprod. Immunol. 35 (2): 101–9. doi:10.1016/s0165-0378(97)00056-9. PMID 9421795.
^ Ober C, Weitkamp LR, Cox N, Dytch H, Kostyu D, Elias S (September 1997). "HLA and mate choice in humans". Am. J. Hum. Genet. 61 (3): 497–504. doi:10.1086/515511. PMC 1715964. PMID 9326314.
^ Chaix R, Chen C, Donnelly P (September 2008). "Is Mate Choice in Humans MHC-Dependent?". PLoS Genetics. 4 (9): e1000184. doi:10.1371/journal.pgen.1000184. PMC 2519788. PMID 18787687.
^ Derti A, Cenik C, Kraft P, Roth FP (April 2010). "Absence of evidence for MHC-dependent mate selection within HapMap populations". PLoS Genetics. 6 (4): e1000925. doi:10.1371/journal.pgen.1000925. PMC 2861700. PMID 20442868.
^ Janeš D, Klun I, Vidan-Jeras B, Jeras M, Kreft S (2010). "Influence of MHC on odour perception of 43 chemicals and body odor". Central European Journal of Biology. 5 (3): 324–330. doi:10.2478/s11535-010-0020-6.
^ Wedekind C, Seebeck T, Bettens F, Paepke AJ (June 1995). "MHC-dependent mate preferences in humans". Proc. Biol. Sci. 260 (1359): 245–9. doi:10.1098/rspb.1995.0087. PMID 7630893.
^ Jacob S, McClintock MK, Zelano B, Ober C (February 2002). "Paternally inherited HLA alleles are associated with women's choice of male odor". Nat. Genet. 30 (2): 175–9. doi:10.1038/ng830. PMID 11799397.
^ Santos, P S; Schinemann JA; Gabardo J; Bicalho MG (April 2005). "New evidence that the MHC influences odor perception in humans: A study with 58 southern Brazilian students". Horm. Behav. 47 (4): 384–388. doi:10.1016/j.yhbeh.2004.11.005. PMID 15777804.
^ "The pill makes women pick bad mates"
^ HLA Alleles Numbers
^ Castro-Prieto A, Wachter B, Sommer S (April 2011). "Cheetah paradigm revisited: MHC diversity in the world's largest free-ranging population". Mol. Biol. Evol. 28 (4): 1455–68. doi:10.1093/molbev/msq330. PMID 21183613.
^ Babik W, Durka W, Radwan J (December 2005). "Sequence diversity of the MHC DRB gene in the Eurasian beaver (Castor fiber)". Mol. Ecol. 14 (14): 4249–57. doi:10.1111/j.1365-294X.2005.02751.x. PMID 16313590.
^ Zhu L, Ruan XD, Ge YF, Wan QH, Fang SG (2007). "Low major histocompatibility complex class II DQA diversity in the Giant Panda (Ailuropoda melanoleuca)". BMC Genet. 8: 29. doi:10.1186/1471-2156-8-29. PMC 1904234. PMID 17555583.
^ Siddle HV, Kreiss A, Eldridge MD, Noonan E, Clarke CJ, Pyecroft S, Woods GM, Belov K (October 2007). "Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial". Proc. Natl. Acad. Sci. U.S.A. 104 (41): 16221–6. doi:10.1073/pnas.0704580104. PMC 1999395. PMID 17911263.
^ Abbas; Lichtman A.H. (2009). "Ch.10 Immune responses against tumors and transplant". Basic Immunology. Functions and disorders of the immune system (3rd ed.). p. A.B. ISBN 978-1-4160-4688-2.
^ Spurgin, Lewis G.; Richardson, David S. (2010-04-07). "How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings". Proceedings of the Royal Society of London B: Biological Sciences. 277 (1684): 979–988. doi:10.1098/rspb.2009.2084. ISSN 0962-8452. PMC 2842774. PMID 20071384.
Bibliography
- Daniel M. Davis, The Compatibility Gene, London, Penguin Books, 2014 (
ISBN 978-0-241-95675-5).
External links
Major+Histocompatibility+Complex at the US National Library of Medicine Medical Subject Headings (MeSH)- Molecular individuality (German online-book 2012)
NetMHC 3.0 server — predicts binding of peptides to a number of different MHC (HLA) alleles- T-cell Group - Cardiff University
- The story of 2YF6: A Chicken MHC
- RCSB Protein Data Bank: Molecule of the Month - Major Histocompatibility Complex
- dbMHC Home, NCBI's database of the Major Histocompatibility Complex