Microarray
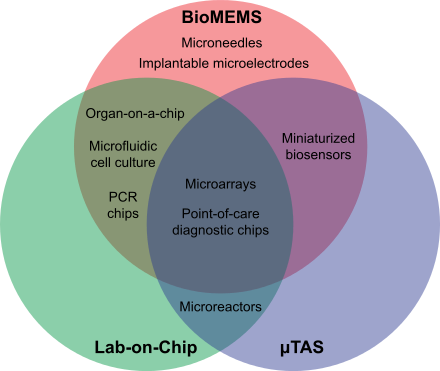
A Venn diagram outlining and contrasting some aspects of the fields of bio-MEMS, lab-on-a-chip, μTAS.
A microarray is a multiplex lab-on-a-chip. It is a two-dimensional array on a solid substrate (usually a glass slide or silicon thin-film cell) that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication[1] and a series of patents.[2] The "gene chip" industry started to grow significantly after the 1995 Science Paper by the Ron Davis and Pat Brown labs at Stanford University.[3] With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophisticated and the most widely used, while the use of protein, peptide and carbohydrate microarrays[4] is expanding.
Types of microarrays include:
DNA microarrays, such as cDNA microarrays, oligonucleotide microarrays, BAC microarrays and SNP microarrays
MMChips, for surveillance of microRNA populations- Protein microarrays
Peptide microarrays, for detailed analyses or optimization of protein–protein interactions
- Tissue microarrays
Cellular microarrays (also called transfection microarrays)- Chemical compound microarrays
- Antibody microarrays
Glycan arrays (carbohydrate arrays)- Phenotype microarrays
Reverse phase protein lysate microarrays, microarrays of lysates or serum- Interferometric reflectance imaging sensor (IRIS)
People in the field of CMOS biotechnology are developing new kinds of microarrays. Once fed magnetic nanoparticles, individual cells can be moved independently and simultaneously on a microarray of magnetic coils. A microarray of nuclear magnetic resonance microcoils is under development.[5]
Fabrication and operation of microarrays
A large number of technologies underlie the microarray platform, including the material substrates, spotting of biomolecular arrays,[6] and the microfluidic packaging of the arrays.[7]
See also
- Microarray databases
- Microarray analysis techniques
Notes
^ Tse-Wen Chang, TW (1983). "Binding of cells to matrixes of distinct antibodies coated on solid surface". Journal of Immunological Methods. 65 (1–2): 217–23. doi:10.1016/0022-1759(83)90318-6. PMID 6606681..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output q{quotes:"""""""'""'"}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ http://www.google.com/patents/US4591570; http://www.google.com/patents/US4829010; http://www.google.com/patents/US5100777.[full citation needed]
^ Schena, M.; Shalon, D.; Davis, R. W.; Brown, P. O. (1995). "Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray". Science. 270 (5235): 467–70. doi:10.1126/science.270.5235.467. PMID 7569999.
^ Wang, D; Carroll, GT; Turro, NJ; Koberstein, JT; Kovác, P; Saksena, R; Adamo, R; Herzenberg, LA; Herzenberg, LA; Steinman, L (2007). "Photogenerated glycan arrays identify immunogenic sugar moieties of Bacillus anthracis exosporium". Proteomics. 7 (2): 180–184. doi:10.1002/pmic.200600478. PMID 17205603.
^ Ham, Donhee; Westervelt, Robert M. (2007). "The silicon that Moves and Feels Small Living Things". IEEE Solid-State Circuits Newsletter. 12 (4): 4–9. doi:10.1109/N-SSC.2007.4785650.
^ Barbulovic-Nad; et al. (2008). "Bio-Microarray Fabrication Techniques—A Review". Critical Reviews in Biotechnology. 26 (4): 237–259. doi:10.1080/07388550600978358.CS1 maint: Explicit use of et al. (link)
^ Zhou; et al. (2017). "Thiol–ene–epoxy thermoset for low-temperature bonding to biofunctionalized microarray surfaces". Lab Chip (21): 3672–3681. doi:10.1039/C7LC00652G.CS1 maint: Explicit use of et al. (link)